Researchers Use Light to Identify Targets for Future Medicines
Hungarian researchers have developed a new photosensitive labelling method to identify the targeted binding of drug molecules to disease-causing proteins. The innovative technology, developed by the Medicinal Chemistry Research Group at the HUN-REN Research Centre for Natural Sciences (HUN-REN RCNS), is not only simpler and more efficient than previous approaches but also more cost-effective and safer. Their findings have drawn the attention of Enamine, one of the world's largest suppliers of research chemicals.
Just as lights once guided ships into unknown harbours, Hungarian researchers are now using specialised light-sensitive molecules to identify new targets for future medicines. Researchers at HUN-REN RCNS have previously studied light-induced protein interactions in detail. Now, they have developed a new technology for securely linking molecules to proteins.
For a drug molecule to be effective, it must modify the function of the disease-causing protein. One way to achieve this is for a drug candidate to bind to a ‘faulty’ protein and disrupt its malfunction. However, a single cell contains tens of thousands of proteins, and molecules must locate suitable binding sites on their surfaces—this is where researchers seek to assist.
To identify the targets of drug molecules, researchers have previously used photosensitive probes that, upon binding, anchor the molecule to a protein cavity when exposed to UV light—much like mooring a ship to the shore with ropes. This anchoring process forms a new chemical bond and alters the mass of the target protein, which can be detected using a specialised instrument called a mass spectrometer.
Led by Péter Ábrányi-Balogh and György Miklós Keserű, researchers at the HUN-REN RCNS Medicinal Chemistry Research Group have now advanced this technology by introducing a new photosensitive probe. Unlike previous solutions, this probe can be easily and inexpensively attached—even as a final step—to an already synthesised drug candidate. The molecule binds only to proteins in the cell that it can match, and upon exposure to light, it marks the exact site of attachment. This new technology not only increases the efficiency of labelling and simplifies the measurement of changes in the target protein’s mass, but also uses UV light close to the visible spectrum, which is less damaging to the target and poses reduced risk to DNA and RNA.
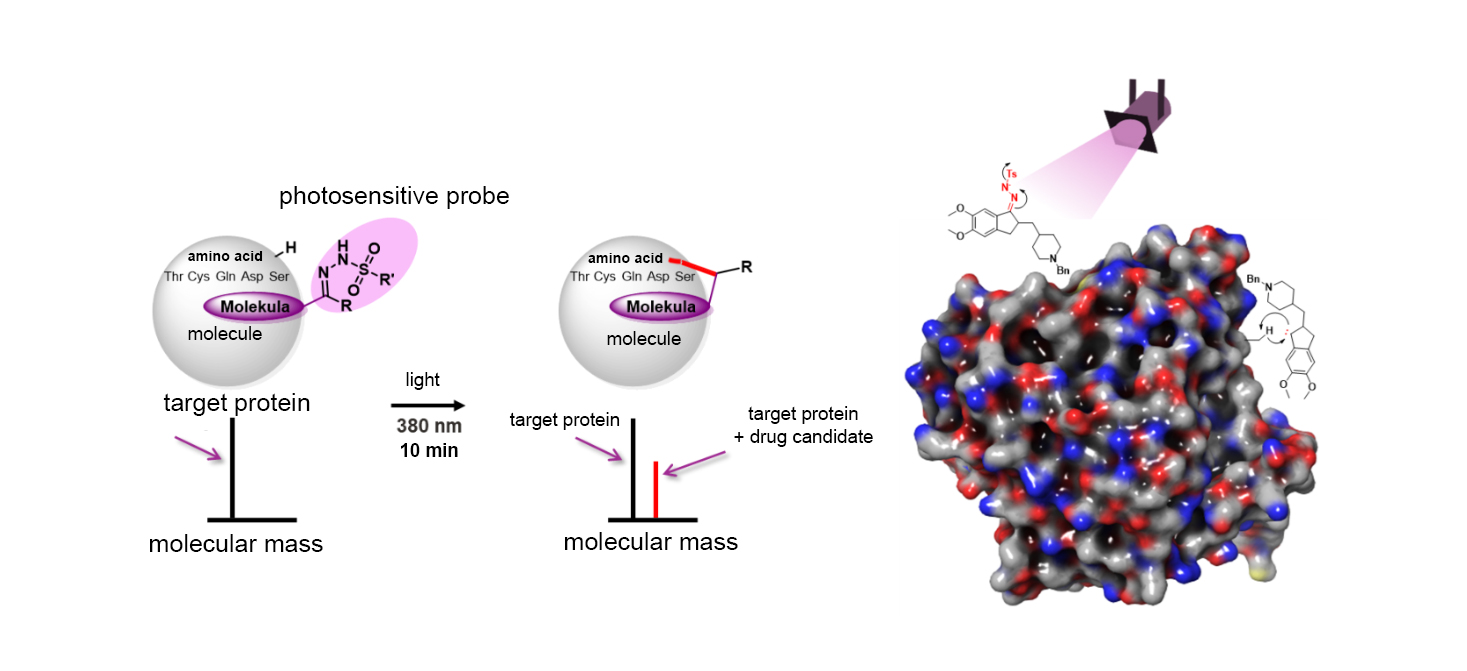
The researchers from HUN-REN RCNS have published their findings in Angewandte Chemie, a leading chemistry journal, demonstrating that the new photosensitive probe can form bonds with a variety of amino acids upon light activation. As a result, drug candidates can be ‘anchored’ to a broad range of proteins, enabling the identification of numerous potential targets.
The broad applicability and efficiency of this method open new avenues for the discovery of drug targets and, consequently, for the development of pharmaceutical treatments for a range of diseases. This approach has been successfully applied to mapping the binding sites of proteins involved in cognitive decline (acetylcholinesterase), depression and other neurological and psychiatric disorders (monoamine oxidase A), and the development of haematological cancers (STAT5 transcription factor). The technique has also proven effective in labelling protein samples derived from cells.
Initial applications have confirmed that this new technology can be used to study drug mechanisms of action and to identify new potential drug targets. These discoveries pave the way for the development of safer and more effective medicines, says György Miklós Keserű, academician and head of the HUN-REN RCNS Medicinal Chemistry Research Group.
The results have attracted the interest of Enamine, the world’s largest supplier of research chemicals, which is now working with the Hungarian researchers to make this new technology more widely accessible to the scientific community. Their aim is to ensure that the necessary reagents are readily available and can be purchased easily and affordably. Looking ahead, the researchers plan to extend the application of this method to label a broad range of biomolecules and to further explore drug mechanisms and potential therapeutic targets.

