The Pathogen Discovery Research Group at the HUN-REN Veterinary Medical Research Institute (HUN-REN VMRI) investigated the virus causing the syndrome associated with reproductive disorders and respiratory symptoms in pigs (PRRS). The sequencing protocol developed by the researchers, which uncovers local strain diversity is expected to help better understand the disease epidemiology and viral evolutionary dynamics, which will contribute to more effective disease control and prevention. The results of the research have been published in the scientific journals Frontiers in Veterinary Science and Animals.
The syndrome associated with reproductive disorders and respiratory symptoms (PRRS) is one of the most economically damaging infectious diseases in pig farming, with estimated global economic damage exceeding USD 2 billion annually. The goal of the National PRRS Eradication Plan (NMT) is to eliminate the infection in the domestic pig population, thereby eliminating all direct and indirect economic damages caused by the virus. In the medium term, the plan aims to reduce antibiotic usage in the pig industry and enhance the international competitiveness of Hungarian pork. One possible way to control the disease is to utilise vaccines. In the defence against PRRS, alongside vaccination, epidemic control measures and elimination of pathogens in the herds play important roles.
The researchers adapted a new, fast, simple and cost-effective PCR-based protocol for next-generation sequencing technology and applied it in two approaches. They analysed the genome stability of the modified live vaccine strain Porcilis MLV employing 34 unique primer pairs, and the ORF7 region of both PRRSV-1 and PRRSV-2 wild-type strains. Using the new method, unique nucleotide variations and length polymorphisms occurring in the genome can be identified. Uncovering local strain diversity is expected to help better understand the disease epidemiology and viral evolutionary dynamics, which will contribute to more effective disease control and prevention.
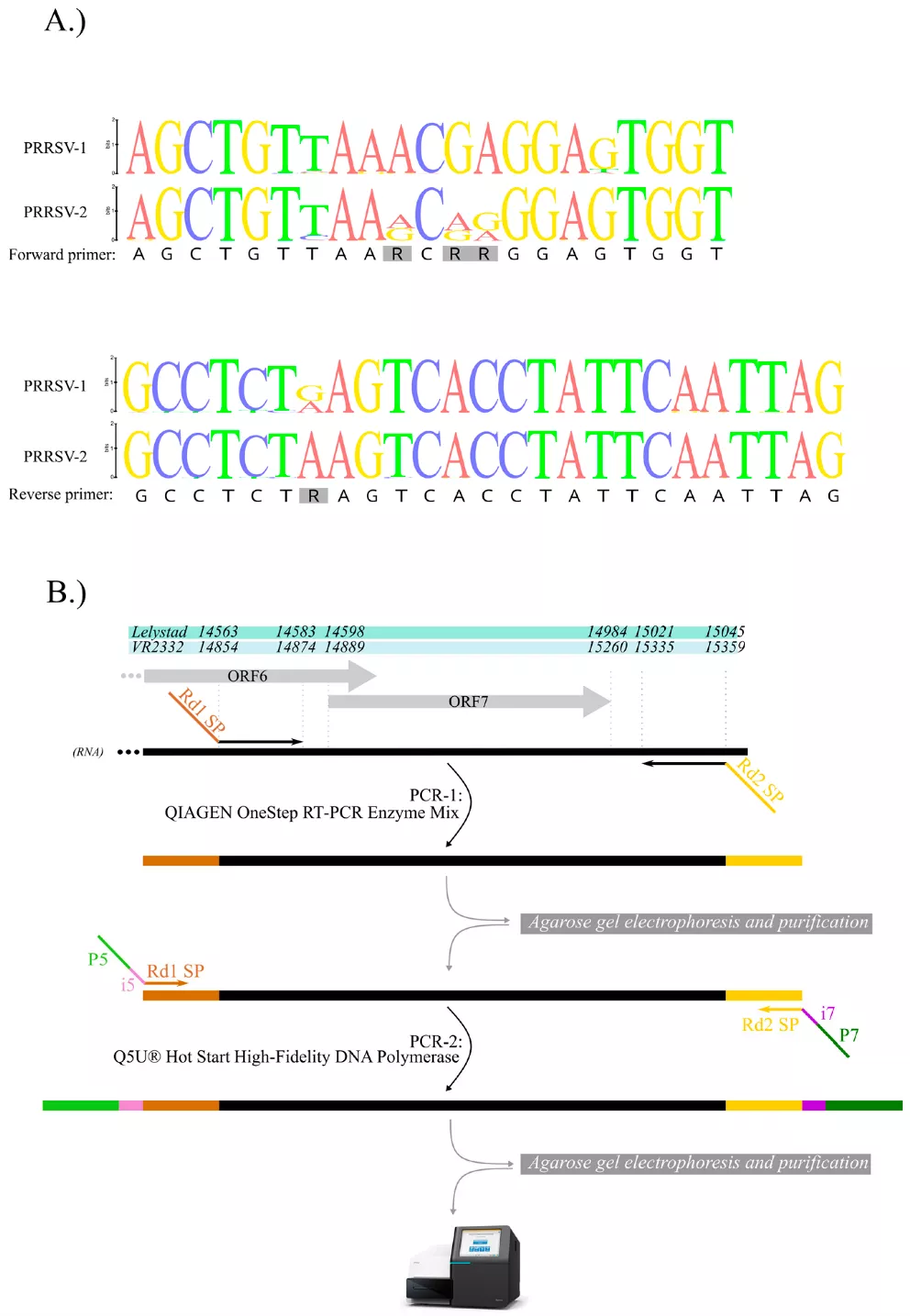
Presentation of a method specific to the ORF7 region. (A) Nucleotide sequence alignment of PRRSV-2 and PRRSV-1 showing the newly designed ORF7-specific primer regions. (B) Position of the amplified ORF7 product on both reference genomes (Lelystad and VR-2332), respectively, and the schematic presentation of the two-step RT-PCR-based library preparation protocol.
The research was conducted within the framework of the "National Laboratory of Infectious Animal Diseases, Antimicrobial Resistance, Veterinary Public Health and Food Chain Safety" project (RRF-2.3.1-21-2022-00001) with support from the Restoration and Resilience Facility / National Restoration Fund, and financed by the RRF-2.3.1-21 grant program.